The model assumes an underlying Poisson point pattern and a Gaussian
response, with intensity
where
Both species are associated through a residuals
Residuals have a multivariate (bivariate) normal distribution with mean zero and covariance matrix,
The inverse of the covariance matrix is called the precision matrix,
denoted by
Function
The function two2tango() needs the following arguments:
mu_1=mu_2=sigma_1=sigma_2=peak_1=peak_2=var=1,cov1=
Then it returns a list with two sf objects of POINT geometry, one
for each species.
library(spatstat)
library(tmap)
tmap_mode("plot")
library(terra)
library(gstat)
library(sf)
library(tidyverse)
# functions
source('code/two2tango.R')
source('code/auxiliary.R')
Test the function
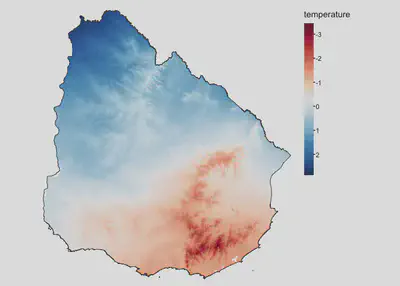
We will use as an example covariate the average annual temperature for Uruguay
uruguay <- geodata::gadm(country = 'UY', level=0, path = 'data/')
temperature <- geodata::worldclim_country('UY', var = 'tavg', path = 'data/')
temperature <- mean(temperature, na.rm=T) %>% mask(uruguay)
temp <- scale(temperature)
tm_shape(temp) +
tm_raster(col.scale = tm_scale_continuous(midpoint = NA, values = 'brewer.rd_bu'),
col.legend = tm_legend('temperature')) +
tm_shape(uruguay) +
tm_borders() +
tm_layout(frame=F, legend.frame = F)
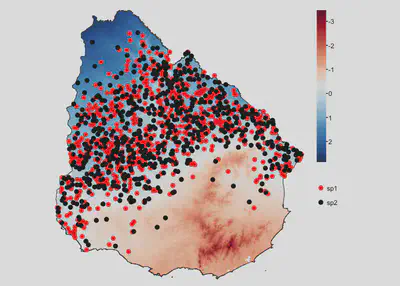
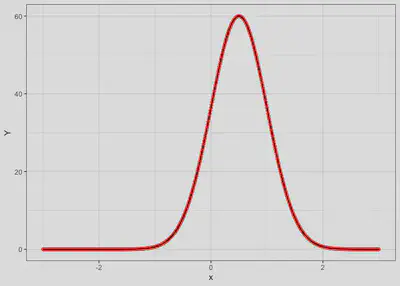
Case 1
Species have the same niche and co-occur
sp1 | 0.5 | 0.5 | 60 | 1 | 0.9 |
sp2 | 0.5 | 0.5 | 60 | 1 | 0.9 |
mu1 = 0.5
mu2 = 0.5
sigma1 = 0.5
sigma2 = 0.5
peak1 = 60
peak2 = 60
cov= 0.9
simulated_species <- two2tango(peak1=peak1, peak2=peak2,
mu1=mu1, sigma1=sigma1,
mu2=mu2, sigma2=sigma2,
cov=cov,
predictor = temp)
sp1 <- simulated_species[[1]] %>% mutate(species = 'sp1')
sp2 <- simulated_species[[2]] %>% mutate(species = 'sp2')
Code
tm_shape(temp) +
tm_raster(col.scale = tm_scale_continuous(midpoint = NA, values = 'brewer.rd_bu'),
col.legend = tm_legend('')) +
tm_shape(uruguay) + tm_borders() +
tm_shape(sp1) +
tm_dots(fill='species',
fill.scale = tm_scale_categorical(values='red'),
fill.legend = tm_legend(''), size = 0.5) +
tm_shape(sp2) +
tm_dots(fill='species',
fill.scale = tm_scale_categorical(values='black'),
fill.legend = tm_legend(''), size = 0.5) +
tm_layout(frame=F, legend.frame = F)
Code
response.df <- tibble(x = seq(-3, 3, by = 0.01),
y1 = spec.response(x, mu1, peak1, sigma1),
y2 = spec.response(x, mu2, peak2, sigma2))
ggplot() +
geom_line(data=response.df, aes(x=x, y=y1), col='red', linetype = 'dashed') +
geom_point(data=response.df, aes(x=x, y=y1), col='red') +
geom_line(data=response.df, aes(x=x, y=y2), col='black') +
geom_line(data=response.df, aes(x=x, y=y2), col='black') +
labs(y='Y') + theme_bw()
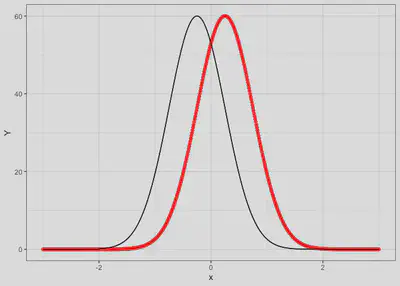
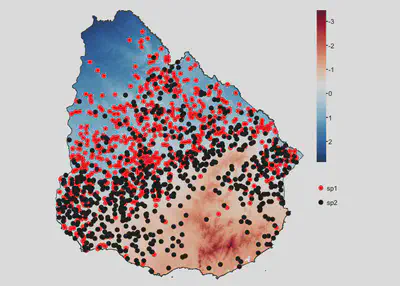
Case 2
Species have a different niche and negative co-occurrence
sp1 | 0.25 | 0.5 | 60 | 1 | -0.9 |
sp2 | -0.25 | 0.5 | 60 | 1 | -0.9 |
mu1 = 0.25
mu2 = -0.25
sigma1 = 0.5
sigma2 = 0.5
peak1 = 60
peak2 = 60
cov= -0.9
simulated_species <- two2tango(peak1=peak1, peak2=peak2,
mu1=mu1, sigma1=sigma1,
mu2=mu2, sigma2=sigma2,
cov=cov,
predictor = temp)
sp1 <- simulated_species[[1]] %>% mutate(species = 'sp1')
sp2 <- simulated_species[[2]] %>% mutate(species = 'sp2')
Code
tm_shape(temp) +
tm_raster(col.scale = tm_scale_continuous(midpoint = NA, values = 'brewer.rd_bu'),
col.legend = tm_legend('')) +
tm_shape(uruguay) + tm_borders() +
tm_shape(sp1) +
tm_dots(fill='species',
fill.scale = tm_scale_categorical(values='red'),
fill.legend = tm_legend(''), size = 0.5) +
tm_shape(sp2) +
tm_dots(fill='species',
fill.scale = tm_scale_categorical(values='black'),
fill.legend = tm_legend(''), size = 0.5) +
tm_layout(frame=F, legend.frame = F)
Code
response.df <- tibble(x = seq(-3, 3, by = 0.01),
y1 = spec.response(x, mu1, peak1, sigma1),
y2 = spec.response(x, mu2, peak2, sigma2))
ggplot() +
geom_line(data=response.df, aes(x=x, y=y1), col='red', linetype = 'dashed') +
geom_point(data=response.df, aes(x=x, y=y1), col='red') +
geom_line(data=response.df, aes(x=x, y=y2), col='black') +
geom_line(data=response.df, aes(x=x, y=y2), col='black') +
labs(y='Y') + theme_bw()